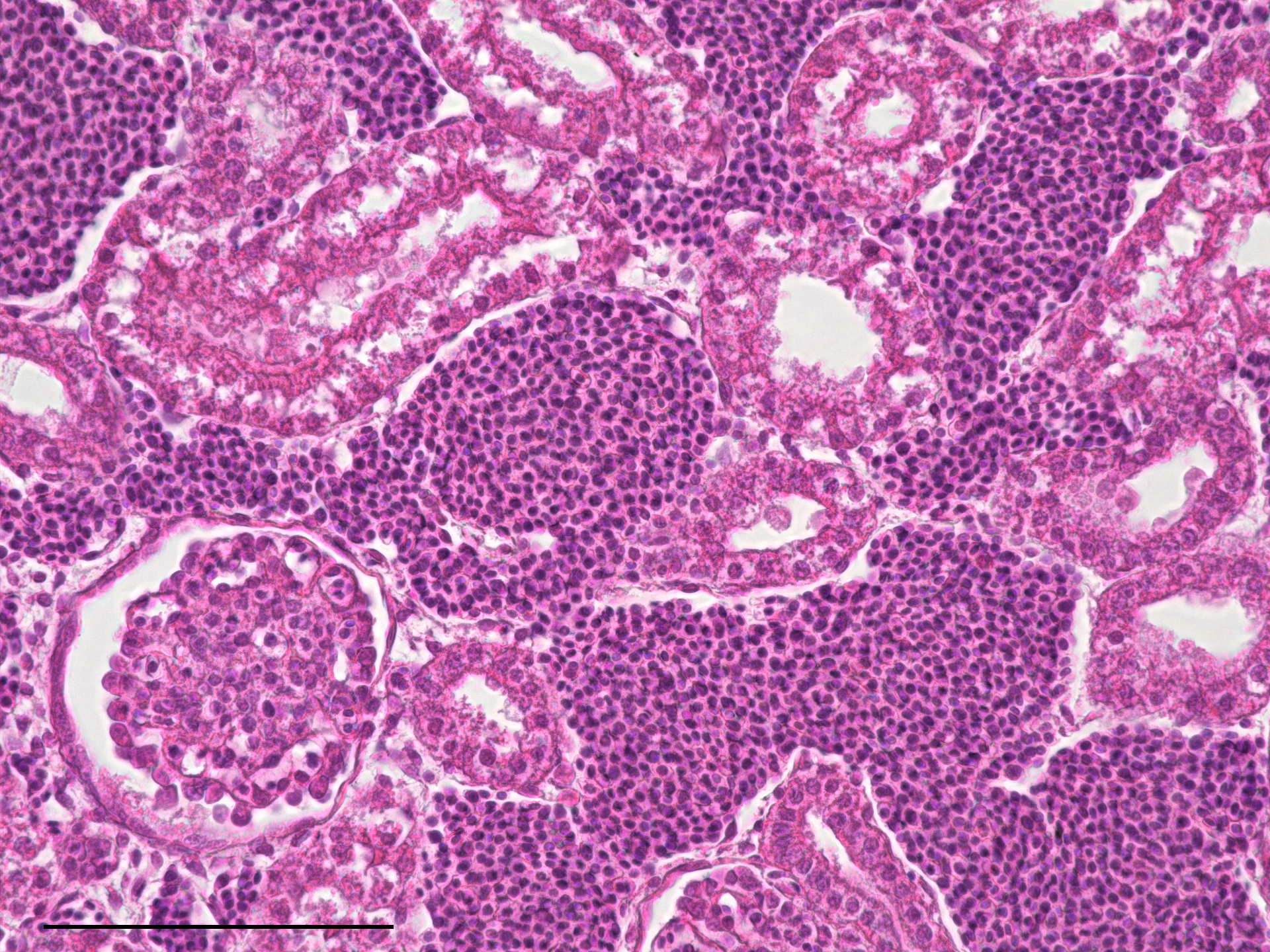
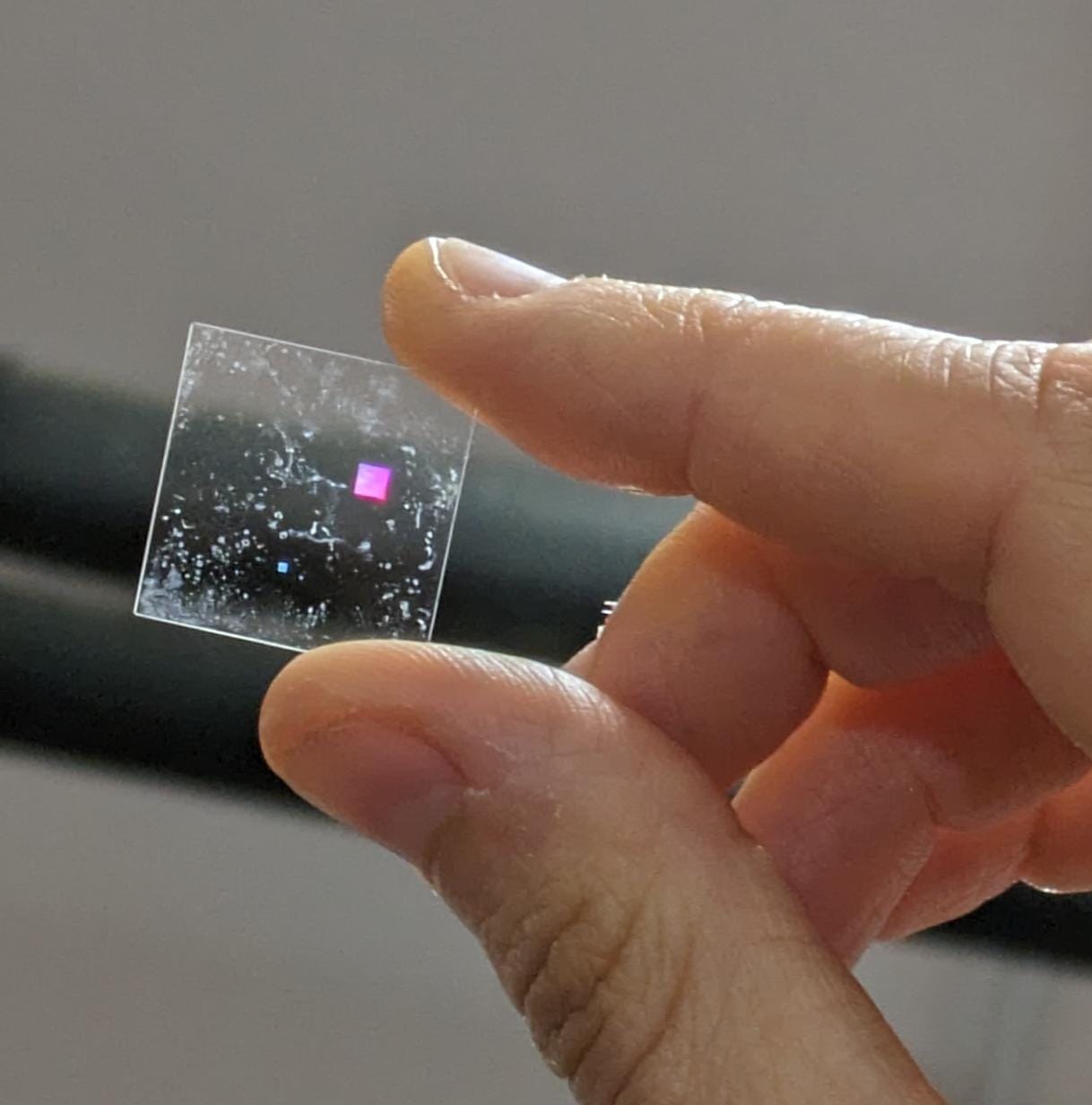
- Design and microfabrication of an implantable device for mesenchymal stem cell maintenance and immunomodulation

This thesis is based at the Mechanobiology lab c/o the Dept. of Chemistry “G. Natta” and the IFN-CNR c/o the Dept. of Physics (Politecnico di Milano – Campus Leonardo). This project aims to create an injectable cellularized device with immunomodulation properties for promoting tissue regeneration. The project will focus on the design and microfabrication of a biocompatible microstructure replicating physiological conditions of Mesenchymal Stem Cells (MSCs)growth and function. Key aspects include designing and developing structures supporting cellular viability and proliferation through optimized porosity, geometry, and integrated nutrient delivery systems and the screening of these samples on the bases of their capability to activate genes related to anti-inflammatory processes. To this aim, advanced microfabrication techniques will be employed to achieve the device’s precise and practical construction. A comprehensive biological validation process will assess the device’s effectiveness in cultivating cells capable of modulating the immune response.
Ideal candidates are students eager to develop knowledge in microfabrication technologies and immune-modulation strategies.
Basic knowledge required: CAD design.
Tutors: Leonardo Cherubin (leonardo.cherubin@polimi.it), Prof. Emanuela Jacchetti (emanuela.jacchetti@polimi.it)
- Antimatter spectroscopy on epithelial cellular models (1 or 2 students). Thesis start: February-March 2025, duration: 10-12 months
This thesis will be conducted at the Mechanobiology Lab (Department of Chemistry, Politecnico di Milano—Campus Leonardo), with opportunities to perform antimatter spectroscopy measurements at the Positron Lab (L-NESS, Department of Physics, Politecnico di Milano—Como).
The project involves a combination of experimental and computational approaches to advance our understanding of tissue behavior in antimatter-based spectroscopic studies. The primary goal is to develop two in vitro models (retinal epithelium and liver tissue) for the evaluation of cells conditions through antimatter interactions. The project will focus on assessing epithelial resistance and permeability in cellular monolayers and comparing untreated cells with treated ones with drugs to alter barrier properties. The student will develop an in-silico model for tissue radiation damage evaluation.
The ideal candidate should be motivated, with a bioengineering background, and open to working in a multidisciplinary research field integrating biology, physics, and computational modeling.
Tutors: Ing. Matteo Vicini (matteo.vicini@polimi.it), Prof. Claudio Conci (claudio.conci@polimi.it), Prof. Rafael Ferragut
- Prototyping and realization of a fluidic system for vascular tissue imaging by antimatter (1 or 2 students). Thesis start: September 2024, duration: 10-12 months
The thesis is based at the Mechanobiology lab. ( Department of Chemistry – Politecnico di Milano – campus Leonardo) with the possibility to perform positron annihilation spectroscopy measurements at the VEPAS lab (Department of Physics – Politecnico di Milano – Como).
The work is experimental and computational. It consists in the optimization and realization of a novel system for interfacing biological matter and accelerated antimatter particles. The candidate will improve the design of a pre-existing device by using CAD softwares (e.g. Inventor, Solidworks) and Multiphysics simulations (e.g. COMSOL). The candidate will be involved also in the prototype realization with additive manufacturing techniques (SLA-DLP) and replica molding, and its functional validation with living cells.
The ideal candidate should have a good knowledge of CAD designing and fluidic transport phenomena.
Tutors: Ing. Matteo Vicini (matteo.vicini@polimi.it), Prof. Claudio Conci (claudio.conci@polimi.it), Prof. Rafael Ferragut, Prof. Manuela T. Raimondi
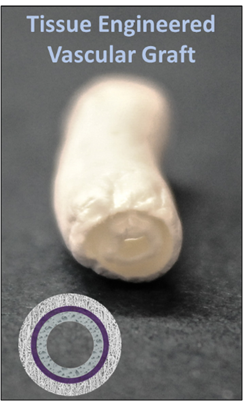
- Cell-Tissue Response in Tissue Engineered Vascular Graft (TEVG)
This thesis is based at the McGowan Institute for Regenerative Medicine (University of Pittsburgh) and at the Bioengineering Department at the University of Pittsburgh (Pittsburgh, USA).
The work is experimental, and it consists in the bio-fabrication of a three-layered vascular grafts (“Multi-Layered Graft for Tissue Engineering Applications,” PCT/US2018/”), decellularization of porcine tissues, cell culture, and in vitro test to investigate cell-tissue specific response in static and dynamic conditions. The candidate will spend 9 months in Pittsburgh (Pittsburgh, USA). Please note that financial funds abroad will not be provided.
The ideal candidate should be motivated for ground-breaking work, interested in tissue engineering, and have a bioing/biotech background. Multicultural/multidisciplinary work attitude is recommended.
Tutors: D’Amore Cardiovascular Tissue Engineering Group (tissueengineering@Fondazionerimed.com), Prof. Claudio Conci (claudio.conci@polimi.it)
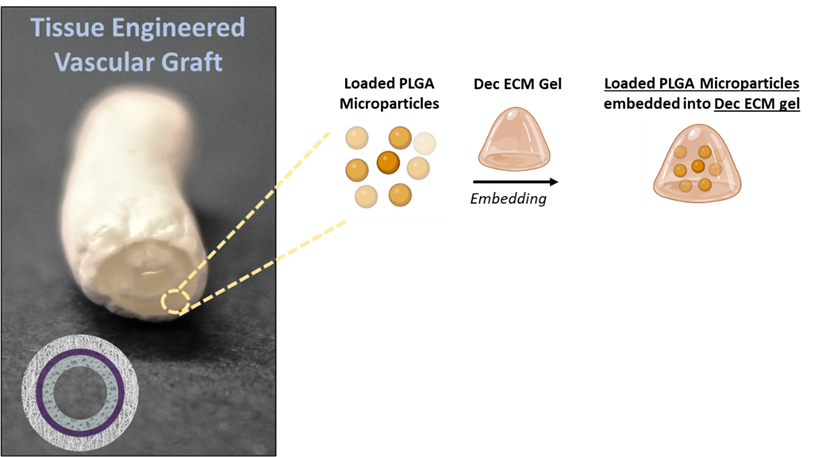
- Anti-inflammatory/bioactive layer to mitigate tissue remodeling in Tissue Engineered Vascular Graft (TEVG)
This thesis is based at the McGowan Institute for Regenerative Medicine (University of Pittsburgh) and at the
Bioengineering Department at the University of Pittsburgh (Pittsburgh, USA). The project is in collaboration with the Department of Pharmacology & Chemical Biology, University of Pittsburgh. The work is experimental and involves some computational tasks. The project consists in the bio-fabrication of a three-layered vascular grafts (“Multi-Layered Graft for Tissue Engineering Applications,” PCT/US2018/”), decellularization of porcine tissues, microparticles fabrication and loading with nitro-oleic acid molecules, characterization of the drug release profile, starting from the release data modelling of the release profile in a 3D structure as the TEVG, in vitro test simulating intimal hyperplasia.
The candidate will spend 9 months in Pittsburgh (Pittsburgh, USA). Please note that financial funds abroad will not be provided. The ideal candidate should be motivated for ground-breaking work, interested in tissue engineering, and have a bioing/biotech background. Multicultural/multidisciplinary work attitude is recommended.
Tutors: D’Amore Cardiovascular Tissue Engineering Group (tissueengineering@Fondazionerimed.com), Prof. Emanuela Jacchetti (emanuela.jacchetti@polimi.it).
- Development of a computational model for transport phenomena in the microvascular environment based on in vitro data
The thesis is based at the Department of Mathematics (MOX) and at the Department of Chemistry (Politecnico di Milano – Campus Leonardo).
The mathematical objective of the thesis is to adapt the model already available to describe the transport phenomena (fluid and transport of chemical substances) in the vascular and extravascular environment to the context of a scaffold implanted in the chicken embryo. The biological objective of the thesis is to understand the interaction between the fibrotic capsule that surrounds the scaffold as its geometric characteristics vary, i.e. how the design of the scaffold influences the microenvironment that forms inside it and how this interacts with the fibrotic capsule that surrounds it.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Prof. Paolo Zunino, Prof. Manuela T. Raimondi
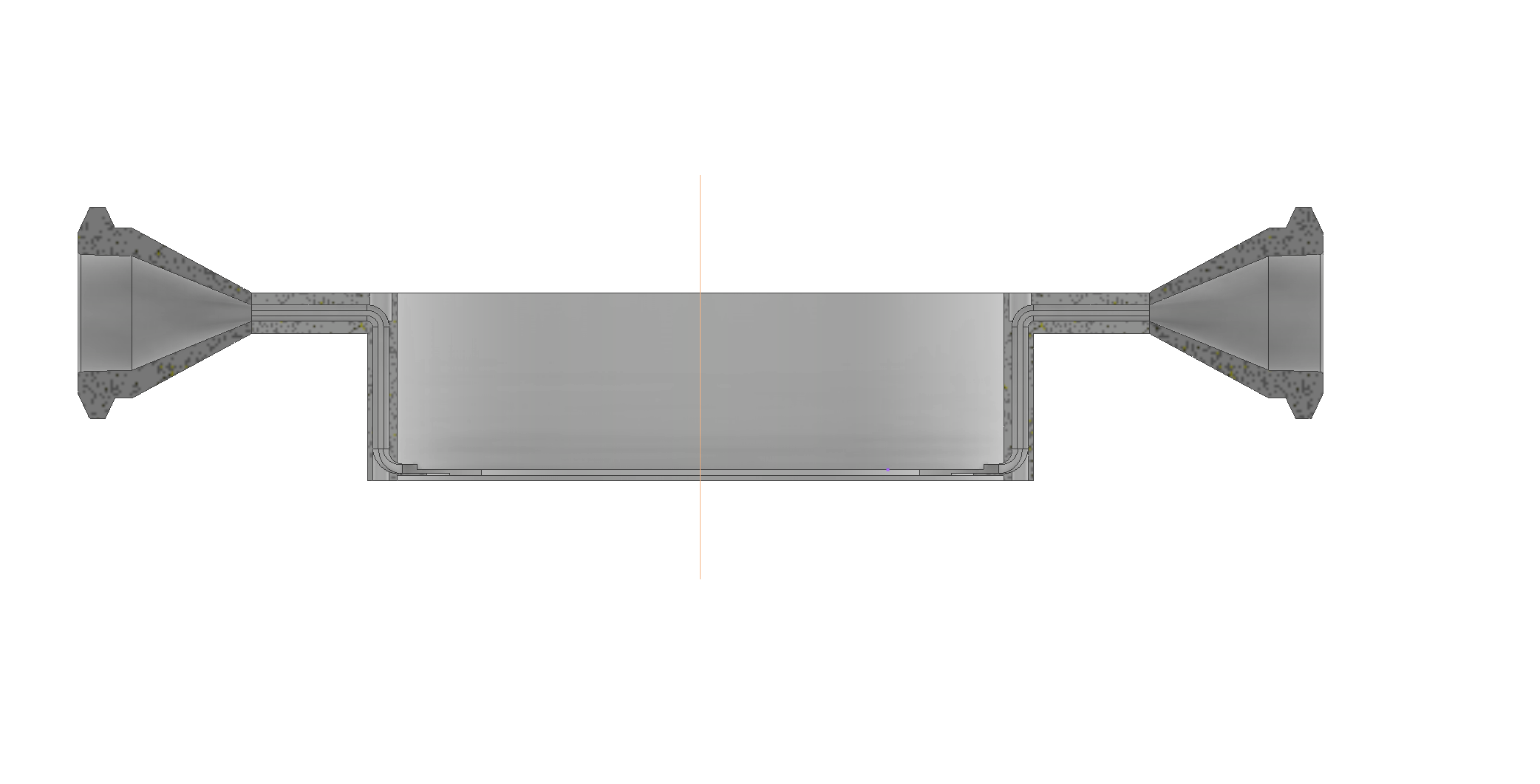
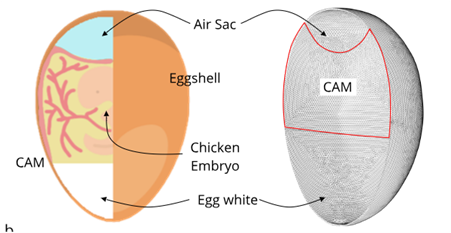
- Design and implementation of a fluidic imaging window for targeted drug delivery in avian embryos. Thesis start: March 2024, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is
computational and experimental.
The work consists in the design, computationa modeling and fabrication of a fluidic bioreactor that will face a living organism. The thesis work will also include: avian embryo cultivation, implantation and microbiological analysis
of a realized chip. Finally, the candidate will quantify fluorescence images acquired by multi-photon acquisition.
The ideal candidate should have a good knowledge of regenerative medicine, Python
programming and capability in 3D modelling.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Leonardo Cherubin (leonardo.cherubin@polimi.it), Prof. Manuela T. Raimondi
- Histological evaluation of breast tumor-mcroenvironment implanted in avian embryo Models: assessing the efficacy of anticancer agents. Thesis start: immediate-start, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is experimental.
The work consists in the implantation of microscaffold realized by two-photon polymerization in avian embryos. The thesis work will include: avian embryo cultivation, implantation and histophatological analysis
of foreign-body reactions. Finally, the candidate will quantify ex-vivo explants by means of histological assays.
The ideal candidate should have a good knowledge of regenerative medicine and biological assays.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Leonardo Cherubin (leonardo.cherubin@polimi.it), Prof. Manuela T. Raimondi
- Synthesis, Polymerization, and Biological Validation of Shape-Memory Resists: Towards the Fabrication of Scaffolds for Tissue Engineering (1 or 2 students). Thesis start: September 2024, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is experimental.
The work consists in the synthesis of a biocompatible photoresist having shape-memory properties. The candidate will test the realized material by two-photon polymerizing microscaffolds. The thesis work will include: material synthesis, additive manufacturing and scanning electron microscopy inspections. Finally, the candidate will test material biocompatibility by standard ISO10993 tests.
The ideal candidate should have a good knowledge of chemistry and additive manufacturing techniques.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Leonardo Cherubin (leonardo.cherubin@polimi.it), Alessandra Nardini (alessandra.nardini@polimi.it), Dr. Rebeca Martinez, Prof. Emanuele Mauri.
- Multicolor photopolymerization of biomcompatible scaffolds for tissue engineering . Thesis start: September 2024, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is experimental.
The work consists in the synthesis of a biocompatible photoresist having the capability to be polymerized with different light-colors. The candidate will test the realized material by two-photon polymerizing microscaffolds. The thesis work will include: material synthesis, additive manufacturing and scanning electron microscopy inspections. Finally, the candidate will test material biocompatibility by standard ISO10993 tests.
The ideal candidate should have a good knowledge of chemistry, additive manufacturing techniques, and light-science.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Alessandra Nardini (alessandra.nardini@polimi.it), Dr. Rebeca Martinez, Prof. Emanuele Mauri
- Microfabrication of permeable glass membrane for oxygen gradient control by in situ molecular diffusion . Thesis start: immediate-start, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is experimental and computational.
The work consists in the microfabrication of microstructured porous glass membranes by femtosecond laser microfabrication techniques. These membranes are designed for the controlled in situ diffusion of oygen/small molecules within cell cultures. The project involves multiple processes, including microfabrication, scanning electron microscopy (SEM), computational simulations, and experimental validation of substrate permeability. Finally, the candidate will biologically validate the substrates through cell culture experiments and particle diffusion assays. Ideal candidates are those interested in advancing their knowledge in laser-based materials technology and biological assays.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Alessandra Nardini (alessandra.nardini@polimi.it), Dr. Rebeca Martinez (rebeca.martinezvazquez@cnr.it), Dr. Chiara Martinelli, Leonardo Cherubin
- Development and characterization of a microtumor environment for anti-cancer drug testing (1 or 2 students). Thesis start: September 2024, duration: 10-12 months
The thesis is based at the Department of Chemistry (Politecnico di Milano – Campus
Leonardo) and at the Department of Physics – CNR-IFN (Politecnico di Milano). The work is
mainly experimental.
The work consists in the microfabrication, cultivation, implantation and microbiological analysis
of a realized microtumor environment on-a-chip. The candidate will culture tumorigenic cell
lines, will characterize the cultivation parameters and will intravital image the in vitro
environment. Finally, the candidate will also implant the chip in living organisms to in vivo test
anti-cancer agents.
The ideal candidate should have a good knowledge of regenerative medicine, Python
programming and capability in 3D modelling.
Tutors: Prof. Claudio Conci (claudio.conci@polimi.it), Leonardo Cherubin (leonardo.cherubin@polimi.it), Dr. Chiara Martinelli (chiara.martinelli@polimi.it), Prof. Manuela T. Raimondi
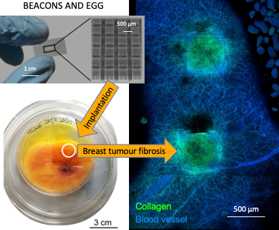
- EGGS&BEACON: bioengineered synthetic eggshell to intravital image living chicken embryos (1 or 2 students). Thesis start: September 2024, duration: 10-12 months
The thesis is based at the Departments of Chemistry (Politecnico di Milano – Campus Leonardo). The work is a challanging computational and experimental experience.
The aim of this project is to overcome principal limitations of the current preclinical models by realizing a new drug testing platform intended as the first union between in vitro and in vivo models. We will develop a radically new platform able to imagine a living organism in real-time in response to angiogenic agents.
The ideal candidate should have a good knowledge of object-oriented coding languages (Python, MATLAB), COMSOL multiphysics (or similar), capability in 3D modelling and a strong attitude in problem solving.
Tutors: Leonardo Cherubin (leonardo.cherubin@polimi.it), Prof. Claudio Conci (claudio.conci@polimi.it), Prof. Manuela T. Raimondi
- Generation of micro-structured 3D millifluidic chips to model endothelial regeneration
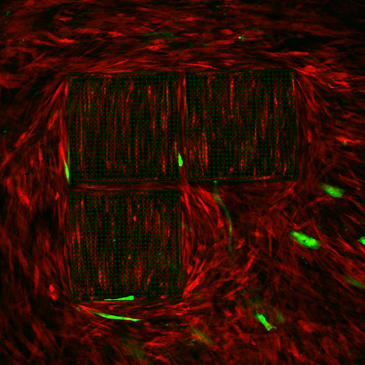

This thesis is based at the Mechanobiology lab c/o the Dept. of Chemistry “G. Natta” and the IFN-CNR c/o the Dept. of Physics (Politecnico di Milano – Campus Leonardo). The work is experimental, and it consists in the use of a millifluidic bioreactor to model endothelial formation. We will analyze the cell adhesion and organization of fibroblast, endothelial cells, and mesenchymal stem cells undergoing mechanical and chemical stimuli (drugs) to monitor tissue (re)generation. Ideal candidates are those willing to develop knowledge in laser-related materials technology and fluorescence microscopy.
Tutors: Dr. Chiara Martinelli (chiara.martinelli@polimi.it), Alberto Bocconi (alberto.bocconi@polimi.it), Prof. Emanuela Jacchetti (emanuela.jacchetti@polimi.it)
- Modeling macrophage phenotype by mechanotrasduction
This thesis is based at the Mechanobiology lab c/o the Dept. of Chemistry “G. Natta” and the IFN-CNR c/o the Dept. of Physics (Politecnico di Milano – Campus Leonardo).
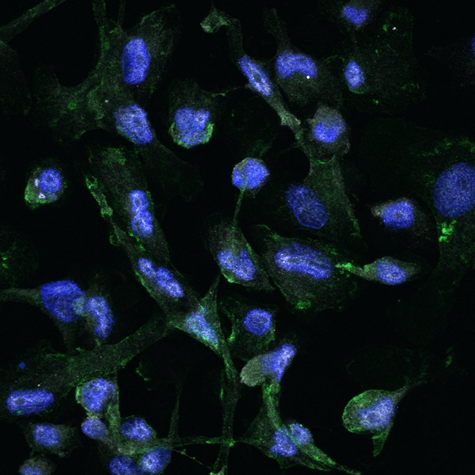

Scaffolds will be mainly fabricated using a micro stereolithography technique called “2-photon laser polymerization”. Cell cultures will be performed using monocyte-derived macrophages, and their polarization will be investigated as a function of different scaffold architectures. Protein expression and localization will be investigated using biochemical assay and high-resolution optical microscopy, such as confocal and FLIM microscopy. Software, like ImageJ and Matlab, will be used to extract informative data from acquired images. Ideal candidates are those willing to develop knowledge in laser-related materials technology and optical microscopy applied to biological systems.
Tutors: Chiara Martinelli (chiara.martinelli@polimi.it), Prof. Emanuela Jacchetti (emanuela.jacchetti@polimi.it)
- Biochemical and mechanical 3D mapping of cancer cells by label-free Brillouin-Raman microspectroscopy
The thesis is based at the Department of Chemistry and at the Department of Physics (Politecnico di Milano – Campus Leonardo). The work is experimental but also involves computational tasks.
To evaluate the role of mechanotransduction in tumor progression and the immune system, the project aim is to study 3D cultured human cancer cells and macrophages mainly using optical
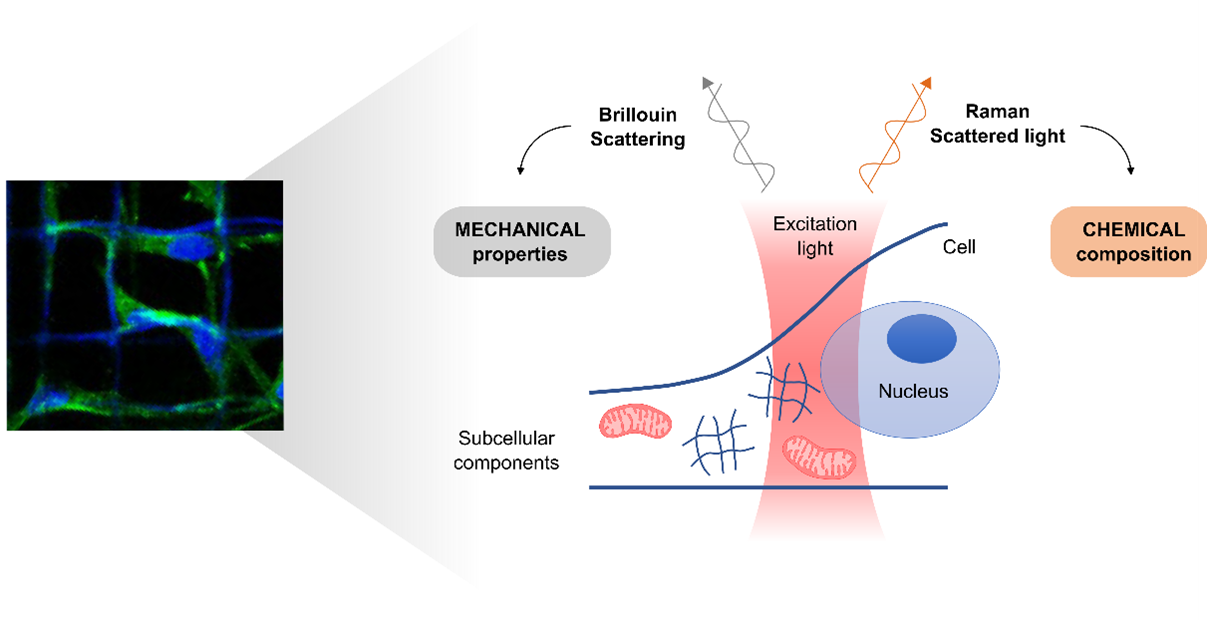

microscopy. The techniques employed will be confocal microscope imaging (to investigate organization and localization of proteins), FLIM imaging (to investigate cell metabolism), Raman imaging (providing detailed biochemical mapping of cells), and Brillouin microscopy (to investigate viscoelastic properties of cells in a non-destructive and contact-free approach).
The candidate will create 3D scaffolds to stimulate cell mechanotransduction and will investigate cell behavior. He/she will optimize the bimodal Raman-Brillouin microscope, and then software like ImageJ and Matlab or programming languages (such as phyton) will be used to extract informative data from acquired images. The project includes a collaboration with a company focused on Brillouin microscopy (Specto photonics).
Tutors and Advisors: Dr. Renzo Vanna (renzo.vanna@cnr.it), Dr. Giuseppe Antonacci (giuseppe@spectophotonics.com), Prof. Dario Polli (dario.polli@polimi.it), Prof. Emanuela Jacchetti (emanuela.jacchetti@polimi.it)
- Implantable medical device design and artificial Intelligence image processing in Foreign Body Response analyses
The thesis is based at the Department of Biophysics (Università di Milano Bicocca – U1) and at the Departments of Chemistry (Politecnico di Milano – Campus Leonardo). The work is mainly computational.
It consists in CAD design of a miniaturized implantable device and its realization with fast
prototyping additive manufacturing techniques (SLA-DLP). The candidate will remotely analyze fluorescence intravital images from mammalian device implantation by coding and applying machine learning algorithms aimed to image restoration. The candidate will use 3D modelling software (e.g. Solidworks, Inventor etc.), MATLAB and Python environments. The ideal candidate should have a good knowledge of object-oriented coding languages (C++, Python, MATLAB), capability in 3D modelling and a strong attitude in problem solving.
Tutors: Ing. Claudio Conci (claudio.conci@polimi.it), Prof. Laura Sironi (laura.sironi@unimib.it), Prof. Manuela T. Raimondi